Single-Cell DNA Sequencing — A Potential Dosimetric Tool
F. Mathew, J. Manalad, L. Galarneau, N. Ybarra, and J. Kildea, and P. N. Tonin, McGill University, Montreal, QC
J. Yeo, National University of Singapore
Y.C. Wang, and I. Ragoussis, McGill Genome Centre, Montreal, QC
Every year, CRPA’s Student and Young Professionals Committee organizes the Anthony J. Mackay Student Paper Contest. The winner receives the Anthony J. MacKay trophy and a $250 cash prize, and the winning paper is published in the CRPA Bulletin. Last year’s winner, Felix Mathew, presented his paper, Single-Cell DNA Sequencing — A Potential Dosimetric Tool, at ICRP 2021+1.
Introduction
It is well known that ionizing radiation (IR) — radiation with enough energy to ionize an atom — can introduce mutations in normal human cells that may transform them into cancer cells over time. It has also been established that different types of radiation have different carcinogenic potentials, though how this affects the integrity of the normal human genome has not yet been characterized. In our research program, we are trying to find a way to investigate and characterize radiation type and dose and their impact on the genomes of radiation-exposed human cells.
In 2016, Behjati et al. reported differences in the mutation patterns of tumours that could differentiate radiation-associated tumours from radiation-naive tumours [1]. Motivated by the Behjati et al. findings, we set out to investigate whether similar mutational information could be used to learn about radiation action in normal human cells. Specifically, we wanted to find out if, by analyzing the mutations induced in vitro in a radiation-exposed group of human cells, we could discern the initiating radiation type and dose. We focused our analyses on two types of radiation: neutrons and photons.
Given that neutrons ionize matter more densely than photons due to their high linear energy transfer (LET), we hypothesized that the mutational patterns of neutrons in exposed cells would be different from those of photons. To test our hypothesis, we investigated mutational effects by single-cell whole genome sequencing (ScWGS) a reference normal human cell line that was exposed to radiation [2].
Methods
This research was carried out in two parts: (1) experimental measurements using human cells, and (2) Monte Carlo simulations of single-cell irradiations to supplement the experimental work.
Experimental measurements
We designed a simple experiment to perform our study as shown in Figure 1. Our plan was to expose human B-lymphoblastoid cells to equal doses of photons and neutrons separately and then compare the induced mutations to evaluate the differences.
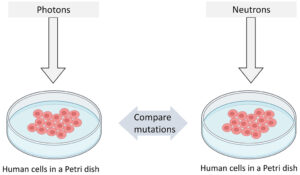
Figure 1: Pictorial representation of our experiment design.
As a first step, we irradiated our cell samples with various doses of 6 MV photons from a medical linear accelerator. Irradiated cells were incubated for 24 hours to allow one DNA damage repair cycle, and subsequently subjected to ScWGS. Large genomic deletion alterations induced by radiation in the genomes of individual cells were identified and quantified.
Monte Carlo simulations
Our group has developed a single-cell nuclear DNA model (shown in Figure 2) [3] using TOPAS-nBio, a Monte Carlo simulation toolkit [4]. To complement our experimental work in part 1, we used this model to computationally examine radiation-induced DNA damage that could potentially lead to the types of large genomic deletion mutations that we can identify in our experimental work.
This DNA model was irradiated with the same doses of radiation (photon or neutron) as in our experiment in part 1. The number of double-strand break (DSB) cluster damages introduced by radiation was measured from the simulations. A DSB cluster refers to a group of DNA damages, within 40 base pairs distance of each other, with a double-strand break also being a part of the group.
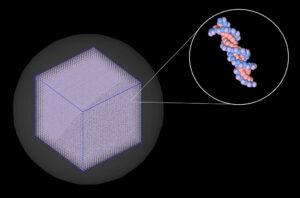
Figure 2: Single-cell nuclear DNA model that we use for Monte Carlo simulations in the TOPAS-nBio simulation toolkit.
Results
In Figure 3, the mean number of large deletion mutations observed in individual cells exposed to 6 MV photons in our experiment is qualitatively compared to the number of DSB clusters measured in cells, as obtained from photon and neutron irradiation simulations. These preliminary experimental results show good agreement with the simulations. Although experimental exposure to neutrons has not yet been undertaken, simulation results confirm our hypothesis that we should expect to see distinct differences between photon- and neutron-induced mutations, at least in terms of dose dependency.
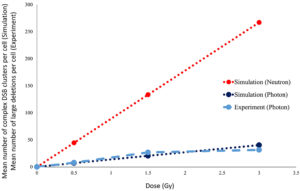
Figure 3: Qualitative comparison of our results from both experiment and simulation of single-cell irradiations with photons and neutrons. To date, the experiment has only been performed with 6 MV photons.
Conclusion
Our preliminary results suggest that ScWGS can be used to quantify the dose dependence of radiation-induced mutations for ionizing photons and neutrons. Our experimental data showed good agreement with our Monte Carlo results, which is promising. However, we have yet to validate our initial findings with repeated experiments. If confirmed, we believe our strategy of examining radiation-induced mutations with single-cell whole-genome sequencing will open up new avenues for radiation biodosimetry.
References
[1] Behjati, S., Gundem, G., Wedge, D. C., Roberts, N. D., Tarpey, P. S., Cooke, S. L., et al. (2016). Mutational signatures of ionizing radiation in second malignancies. Nature communications, 7, 12605. https://doi.org/10.1038/ncomms12605
[2] Li, R., Couturier, C., Savage, P., Monlong, J., Bourque, G., Petrecca, K., Park, M., & Ragoussis, J. (2018). Abstract 2177: Sensitive single cell copy number profiling using a novel microfluidic droplet based platform. Cancer Research, 78(13_Supplement), 2177–2177. https://doi.org/10.1158/1538-7445.am2018-2177
[3] Montgomery, L., Lund, C. M., Landry, A., & Kildea, J. (2021). Towards the characterization of neutron carcinogenesis through direct action simulations of clustered DNA damage. Physics in Medicine & Biology, 66(20), 205011. https://doi.org/10.1088/1361-6560/ac2998
[4] Schuemann, J., McNamara, A. L., Ramos-Méndez, J., Perl, J., Held, K. D., Paganetti, H., et al. (2018). Topas-nBio: An extension to the TOPAS Simulation Toolkit for cellular and sub-cellular radiobiology. Radiation Research, 191(2), 125. https://doi.org/10.1667/rr15226.1
 Felix Mathew
Felix Mathew
Felix is a PhD student in the Biological and Biomedical Engineering department of McGill University. His research is trying to determine mutation signatures of high-LET radiation, particularly neutrons, through single-cell whole-genome sequencing as part of Dr. John Kildea’s Neutron-Induced Carcinogenic Effects research team as a graduate student in the medical radiation physics program.
Do you want to read more articles like this?
The Bulletin is published by the Canadian Radiation Protection Association (CRPA). It’s a must-read publication for radiation protection professionals in Canada. The editorial content delivers the insights, information, advice, and valuable solutions that radiation protection professionals need to stay at the forefront of their profession.
Sign up today and we’ll send you an email each time a new edition goes live. In between issues, check back often for updates and new articles.
Don’t miss an issue. Subscribe now!
Subscribe


 Felix Mathew
Felix Mathew
